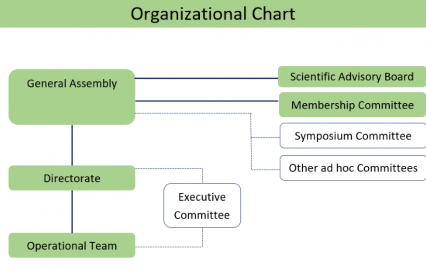
Organization
The WIN Consortium is organized in the following groups who collectively work together to achieve WIN's common goals
“New Leadership, vision for the Worldwide Innovative Network Consortium in Precision Oncology” View Summary
WIN was formed on the premise that we can accomplish more together than each organization can achieve working alone. We aim to improve cancer patients’ survival and quality of life. View WIN's history and unique attributes:
WIN members collaboratively design and carry out global studies designed to achieve breakthroughs for patients worldwide. Our distinguished Scientific Advisory Board oversees WIN studies. Current trials include:
WIN leaders are selected for their contributions and commitment to making effective, personalized cancer medicine a reality for patients around the world. They guide WIN's strategic, operational, and scientific direction.

The WIN Consortium is organized in the following groups who collectively work together to achieve WIN's common goals

Chair, WIN Consortium; Director, Legorreta Cancer Center at Brown University (USA)

Chair Emeritus, WIN Consortium; Professor Emeritus, University of Chicago; past President and former Executive Vice President & Chief Medical Officer (ASCO) (USA)

Chief Medical Officer, WIN Consortium; Prof. of Medicine, Associate Director Clinical Research, MCW Cancer Center and Linda T. & John A. Mellowes Chair of Precision Oncology (USA)

Director, Operational Team, WIN Consortium
WIN members include 34 leading organizations representing all stakeholders in personalized cancer medicine covering 18 countries and 5 continents. Our shared vision is delivering the promise of effective, personalized cancer medicine to patients worldwide. The WIN Consortium has collaborated with additional academic institutions and pharmaceutical partners in successful completion of global clinical trials in precision oncology.
WIN Symposia, held annually, gathers leaders representing a breadth of stakeholders from around the world to learn, share, and collaborate. Our latest symposium took place in Abu Dhabi, UAE, for further information, visit: win-burjeel-symposium.com/

Abstracts Presented at the 2024 WIN Symposium in Partnership with Burjeel Holdings March 1-2, 2024, Abu Dhabi, United Arab Emirates
March 01, 2024 - March 02, 2024The WIN Symposium 2024, a landmark event in precision oncology, took place in Abu Dhabi, United Arab Emirates (UAE), on the 1st and 2nd of March 2024; marking a significant collaboration between the Worldwide Innovative Network (WIN) Consortium and Burjeel Holdings. This annual global congress of the WIN Consortium brought together physicians, researchers, and scientists from 30 countries to explore the latest advancements in precision oncology, with the ultimate goal of improving cancer patient care and outcomes worldwide.
The call for abstracts attracted 51 submissions with 42 accepted as posters and 3 as oral presentations. This publication features 34 of the accepted abstracts.
The abstracts are now published online on the JIPO website:
https://meridian.allenpress.com/innovationsjournals-JIPO/article/doi/10.36401/JIPO-24-X2/500828/Abstracts-Presented-at-the-2024-WIN-Symposium-in
or can be downloaded from this section.

WIN 2024 Symposium
March 01, 2024 - March 02, 2024The two-day WIN Symposium 2024, organized by the WIN Consortium and hosted by Burjeel Holdings took place in Abu Dhabi, United Arab Emirates on 1-2 March 2024.
Under the theme of “Precision and Molecular Oncology: Caring for Patients and Future Generations,” the symposium brought together more than 450 expert clinicians, researchers and industry professionals from over 30 countries. Together, they explored the latest advances in precision oncology and offered insights into the developments shaping the future of oncology. The symposium started with an inspiring keynote lecture by Nobel Laureate Professor James Allison, who has revolutionized cancer immunotherapy. The curated programs featured interactive elements illustrating precision cancer treatment principles and best practices.
More than 50 abstracts were received from over 15 countries. The event was endorsed by the American Society of Clinical Oncology (ASCO®) and accredited for 8.5 European CME credits (ECMEC®s) by the European Accreditation Council for Continuing Medical Education (EACCME®).
The WIN Symposium provided attendees with a deep understanding of the role of key biomarkers, their optimal application, and emerging new omic tests that will inform future biomarker adoption under the two big umbrellas of Precision Genomics and Precision Immunotherapy. Local experience from the UAE, the host country, was included to link global evidence to regional practice. Application of the principles of precision medicine were discussed beyond medical and onto radiation oncology along with issues of access, equity and diversity in precision oncology practice and trials on a global scale. The symposium featured a Molecular Tumor Board to exemplify with real patient cases how to use biomarkers to inform therapeutic choices. Finally, a panel discussion was dedicated to navigating available diagnostic pathways and tests from bench to bedside.
The symposium provided networking opportunities, allowing participants to connect with peers, establish collaborations, and forge new partnerships.
Visit our dedicated symposium website for all the details of this event: https://win-burjeel-symposium.com/.
ASCO® is a registered trademark of the American Society of Clinical Oncology®. Used with permission. This is not an ASCO sponsored event.

WIN 2022 Symposium
October 29, 2022 - October 30, 2022The WIN 2022 Symposium ‘Integrating Genomics and Transcriptomics to Reshape Precision Oncology: A WINning Strategy’ took place on 29-30 October, 2022 in Barcelona, Spain.
We had an exciting line-up of prominent speakers and panel discussion and our 1st international molecular board!